In Silico Study: Molecular Docking and Toxicity Prediction of Pyrazoline Derivatives with Potential as Anti-Inflammatory
DOI:
https://doi.org/10.55749/ijcs.v4i2.73Keywords:
Anti-Inflammatory, Molecular docking, Pyrazoline, ToxicityAbstract
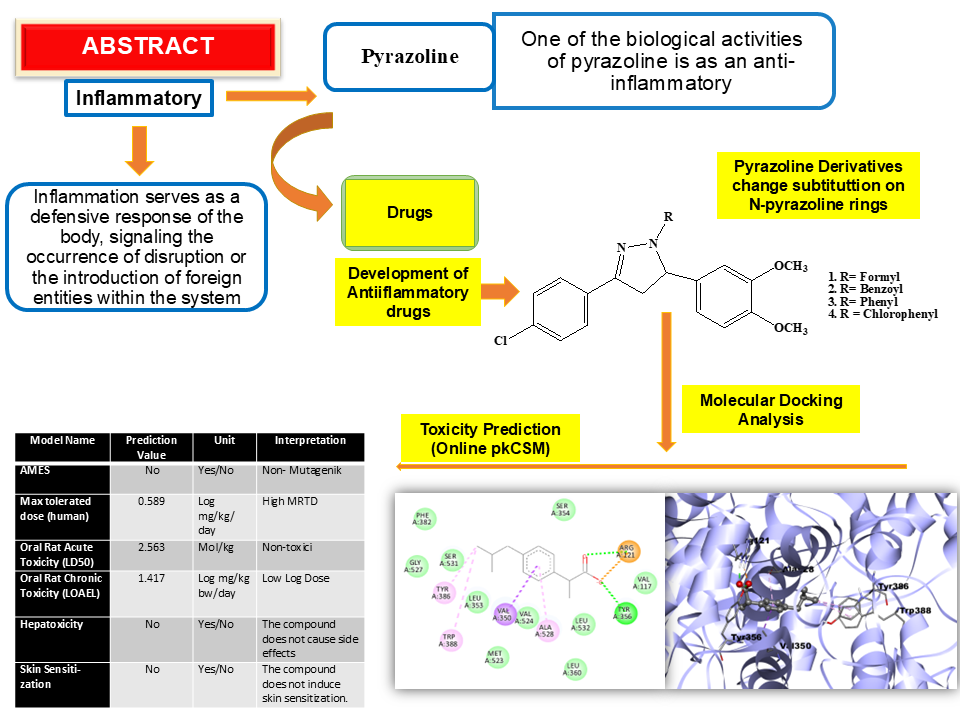
Research on heterocyclic compounds suggested that pharmacologically active agents featuring pyrazoline played a crucial role in medicinal chemistry. When fused with other heterocycles, pyrazoline, as a quiescent heterocyclic moiety, resulted in the enhancement of biological properties. Therefore, synthesizing these compounds had attracted the attention of researchers focused on designing novel drugs. The addition of substituents to the N-pyrazoline atom and modifications to the benzene ring of pyrazoline compounds were essential for the identification of pyrazoline derivatives exhibiting enhanced biological activity. Extensive research had shown that pyrazoline compounds had significant biological effects, including anti-inflammatory effects. Inflammation was the body's reaction to infection or injury and marked by symptoms such as redness, heat, swelling, and pain. This research involved a computational analysis of pyrazoline compounds utilizing molecular docking with AutoDock Tools and AutoDock Vina software on four pyrazoline derivative compounds (pyrazolines 1-4). Simultaneously, their toxicity was assessed through online pkCSM to evaluate their potential as anti-inflammatory drug candidates. The interaction between the active site of cyclooxygenase-2 (COX-2) receptor (PDB: 4PH9) and pyrazoline derivatives showed that pyrazoline 2 (1-benzoyl-(3-(4-chlorophenyl)-5-(3,4-dimethoxy)-4,5-dihydro-2-pyrazoline) exhibited the highest binding affinity of -8.0 kcal/mol compared to pyrazoline derivatives 1, 3, 4 and ibuprofen as native ligands also in the molecular docking test with values of -7.1; -7.7; -7.6; and -7.1 kcal/mol, respectively. Toxicity evaluation for pyrazoline 2 also suggested that this compound was non-toxic, non-hepatotoxic, and did not induce skin sensitization, with an Oral Rat Acute Toxicity (LOAEL) score of 1.417 log (mg/kg_bw/day).
References
[1] Peerzade, N.A., Jadhav, S.Y., & Bhosale, R.B. 2020. Synthesis, molecular docking and biological evaluation of some novel n-carbamoyl pyrazolines as potent anti-inflammatory, antioxidant and antidiabetic agents. Rasayan J. Chem. 13(3). 1401–1411. doi: https://doi.org/10.31788/RJC.2020.1335637.
[2] Nattava, P.L. 2023. Docking studies of indole assimilated pyrazoline molecular hybrids: design, synthesis as antiinflammatory agents and anticancer agents. J. Adv. Sci. Res. 14(07). 22–31. doi: https://doi.org/10.55218/jasr.202314704.
[3] Sahu, S., Banerjee, M., Samantray, A., Behera, C., MA, A., & Azam. 2008. Synthesis, analgesic, anti-inflammatory and antimicrobial activities of some novel pyrazoline derivatives. Trop. J. Pharm. Res. 7(2). 961–968. doi: https://doi.org/10.4314/tjpr.v7i2.14664.
[4] Vasudha, D., Jagadeesh, A., Mali, S.N., Bhandare, R.R., & Shaik, A.B. 2024. Synthesis, molecular docking and pharmacological evaluations of novel naphthalene-pyrazoline hybrids as new orally active anti-inflammatory agents. Chem. Phys. Impact. 8. 100500. doi: https://doi.org/10.1016/j.chphi.2024.100500.
[5] Mohammed K,M., & Omar, T.N.-A. 2023. Synthesis, characterization, anti-inflammatory, and antimicrobial evaluation of new 2-pyrazolines derivatives derived from guaiacol. Iraqi J Pharm Sci. 32(2). 254–261. doi: https://doi.org/10.31351/vol32issSuppl.pp254-261.
[6] Al-Warhi, T. et al. 2022. Development of novel isatin thiazolyl-pyrazoline hybrids as promising antimicrobials in MDR pathogens. RSC Adv. 12(48). 31466–31477. doi: https://doi.org/10.1039/d2ra04385h.
[7] Fadhil, H.R., Raauf, A.M.R., & Mahdi, M.F. 2024. Synthesis, characterization, preliminary molecular docking, pharmacological activity, and ADME studies of some new pyrazoline derivatives as anti-breast cancer agents. Pharmacia. 71. 1–10. doi: https://doi.org/10.3897/pharmacia.71.e133015.
[8] Mazyed, H.A., Razzaq, A.S., Hussein, G.H., & Nahi, R.J. 2022. Synthesis and anti-diabetic activity evaluation of new 1,2,3-triazole derivatives incorporating 2-pyrazoline ring. Int. J. Health Sci. (Qassim). 6(May). 7299–7307. doi: https://doi.org/10.53730/ijhs.v6ns4.10171.
[9] Karrouchi, K., Radi, S., Ramli, Y., Taoufik, J., Mabkhot, Y.N., Al-Aizari, F.A., & Ansar, M. 2018. Synthesis and pharmacological activities of Pyrazole derivatives: A review, 23(1). doi: https://doi.org/10.3390/molecules23010134.
[10] Bhadoriya, U. & Jain, D. 2018. Synthesis, characterization, anti inflammatory activity and molecular docking study of novel pyrazoline derivatives bearing chalcone backbone. Indian J. Chem. -Section B. 57(10). 1315–1321.
[11] Ahsana, D., Andika, A., & Nashihah, S. 2021. Molecular Docking Study of Flavonoid Compounds in The Guava Leaves (Psidium Guajava L.) Which Has Potential as Anti-Inflammatory COX-2 Inhibitors. Lumbung Farm. J. Ilmu Kefarmasian. 2(2). 67. doi: https://doi.org/10.31764/lf.v2i2.5487.
[12] Ma, X. 2022. Development of Computational Chemistry and Application of Computational Methods. J. Phys. Conf. Ser. 2386(1). doi: https://doi.org/10.1088/1742-6596/2386/1/012005.
[13] Wiratama, M. & Budimarwanti, C. 2025. Synthesis and Molecular Docking Study of Dibenzal Monocarbonyl ( Curcumin Analog ) and Its Potential as Anti-Inflammatory. J. Sci. Appl. Chem. 28(2). 68–72. doi: https://doi.org/10.14710/jksa.28.2.68-72.
[14] Agu, P.C., Afiukwa, C.A., Orji, O.U., Ezeh, E.M., Ofoke, I.H., Ogbu, C.O., Ugwuja, E.I., & Aja, P.M. 2023. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 13(1). 1–18. doi: https://doi.org/10.1038/s41598-023-40160-2.
[15] Castro-Alvarez, A., Costa, A.M., & Vilarrasa, J. 2017. The Performance of several docking programs at reproducing protein-macrolide-like crystal structures. Molecules. 22(1). doi: https://doi.org/10.3390/molecules22010136.
[16] Wira Waskitha, S.S., Istyastono, E.P., & Octa Riswanto, F.D. 2023. Molecular Docking Study of Caffeic Acid as Acetylcholinesterase Inhibitor. J. Food Pharm. Sci. 11(3). 867–873. doi: https://doi.org/10.22146/jfps.7665.
[17] Wiratama, M., Satria, S., Waskitha, W., Haryadi, W., & Wahyuningsih, T.D. 2022. Synthesis, antimalarial activity assay and molecular docking study of N-substituted chloro-pyrazolines. Trop. J. Pharm. Res. 21(6). 1255–1261. doi: https://doi.org/10.4314/tjpr.v21i6.18.
[18] Pampalakis, G. 2023. Underestimations in the In Silico-Predicted Toxicities of V-Agents. J. Xenobiotics. 13(4). 615–624. doi: https://doi.org/10.3390/jox13040039.
[19] Pires, D.E.V., Blundell, T.L., & Ascher, D.B. 2015. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 58(9). 4066–4072. doi: https://doi.org/10.1021/acs.jmedchem.5b00104.
[20] Bursch, M., Mewes, J.-M., Hansen, A., & Grimme, S. 2022. Best-Practice DFT Protocols for Basic Molecular Computational Chemistry, Angewandte Chemie. 61. doi: https://doi.org/10.1002/anie.202205735.
[21] Govindarajan, M., Abdelhameed, A.S., Al-Saadi, A.A., & Attia, M.I. 2015. Experimental and theoretical studies of the vibrational and electronic properties of (2E)-2-[3-(1H-imidazol-1-yl)-1-phenylpropylidene]-N-phenylhydrazinecarboxamide: An anticonvulsant agent. Appl. Sci. 5(4). 955–972. doi: https://doi.org/10.3390/app5040955.
[22] Ramírez, D. & Caballero, J. 2018. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules. 23(5). 1–17. doi: https://doi.org/10.3390/molecules23051038.
[23] R. Bharathi. N. Santhi. 2020. In vitro and molecular docking studies of an antiinflammatory scaffold with human peroxiredoxin 5 and tyrosine kinase receptor. Bioinformation. 16(11). 929–936. doi: https://doi.org/10.6026/97320630016923.
[24] Thomas, D.N., Wills, J.W., Tracey, H., Baldwin, S.J., Burman, M., Williams, A.N., Harte, D.S.G., Buckley, R.A., & Lynch, A.M. 2024. Ames test study designs for nitrosamine mutagenicity testing: qualitative and quantitative analysis of key assay parameters. Mutagenesis. 39(2). 78–95. doi: https://doi.org/10.1093/mutage/gead033.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Indonesian Journal of Chemical Studies

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.