Novel Natural Deep Eutectic Solvent (NaDES) Yellow Choline Chloride and Molecular Docking Soybean Extract (Glycine max) as Diabetes Drugs Candidate
DOI:
https://doi.org/10.55749/ijcs.v3i2.47Keywords:
Glycine max, Molecular Docking, Natural Deep Eutectic Solvent (NaDES), UV-Vis spectrophotometer, Yellow Choline ChlorideAbstract
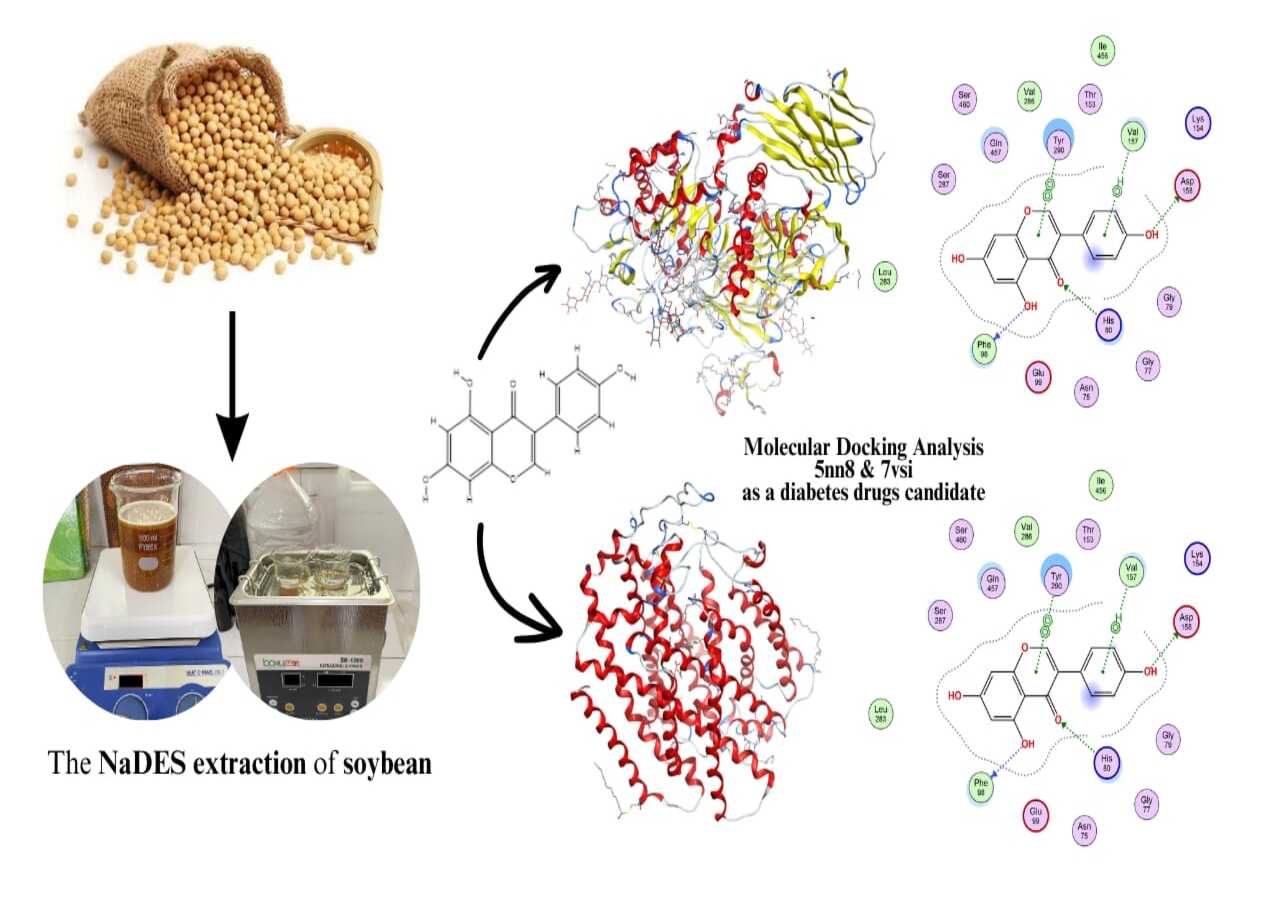
Natural Deep Eutectic Solvent (NaDES) is an environmentally friendly extraction method to obtain soybean bioactive compounds, focusing on genistein compounds as drug candidates. The use of environmentally friendly extraction solvents could support green extraction to ensure the safety of natural medicinal candidates. HBA (Hydrogen Bonding Acceptor), yellow choline chloride (supplement in animal feed), and HBD (Hydrogen Bonding Donor) lactic acid. A UV-Vis spectrophotometer was used to detect genistein. MoE 2022.02 software was used in the molecular docking simulation, and the docking scoring methods affinity ΔG and GBVI/WSA (induced fit) were used. The PDB ID used was: 5nn8 (alpha glucosidase) and PDB ID: 7vsi (SGLT-2 Inhibitor). The results of genistein were obtained by 92,670 mg (0.9267%) in the 75 0C, 30 min ultrasonic NaDES extraction in HBD lactic acid. Genistein exhibited an affinity for the 5NN8 (alpha-glucosidase) and 7VSI (SGLT-2 Inhibitor) receptors of -6,230 and -8,768, respectively. These affinity values did not exceed the interaction values of the native ligands acarbose (alpha-glucosidase) and Empagliflozin (SGLT-2 Inhibitor), which were -8,988 and -12,302, respectively. Genistein compounds had the lowest RMSD value of 0.819 at 7vsi (SGLT-2 Inhibitor). These results suggested the possibility of a genistein pathway as a candicate diabetes drug. The NaDES extraction method demonstrated great potential for development into a green action that supported the green extraction process, and genistein was an isoflavone compound that could be a candidate for diabetes drugs.
References
Oomen W.W., Begines P., Mustafa N. R., Wilson E. G., Verpoorte R., & Choi Y.H. 2020. Natural deep eutectic solvent extraction of flavonoids of Scutellaria baicalensis as a replacement for conventional organic solvents. Molecules. 25(3). 617. doi: https://doi.org/10.3390/molecules25030617.
Syarifah A.N., Suryadi H., Hayun H., Simamora A., & Mun’im A. 2023. Detoxification of comfrey (Symphytum officinale L.) extract using natural deep eutectic solvent (NADES) and evaluation of its anti-inflammatory, antioxidant, and hepatoprotective properties. Front. Pharmacol. 14. 1012716. doi: https://doi.org/10.3389/fphar.2023.1012716.
Yang G.Y., Song J.N., Chang Y.Q., Wang L., Zheng Y.G., Zhang D., & Guo L. 2021. Natural deep eutectic solvents for the extraction of bioactive steroidal saponins from Dioscoreae nipponicae rhizome. Molecules. 26(7). 2079. doi: https://doi.org/10.3390/molecules26072079.
Martinović M., Krgović N., Nešić I., Žugić A., & Tadić V.M. 2022. Conventional vs. green extraction using natural deep eutectic solvents—Differences in the composition of soluble unbound phenolic compounds and antioxidant activity.Antioxidants. 11(11). 2295. doi: https://doi.org/10.3390/antiox11112295.
Lupidi G., Palmieri A., & Petrini M. 2022. Sustainable and fast synthesis of functionalized quinoxalines promoted by natural deep eutectic solvents (NADESs). Green Chem. 24(9). 3629-3633. doi: https://doi.org/10.1039/D2GC00664B.
Ivanović M., Islamčević Razboršek M., & Kolar M. 2020. Innovative extraction techniques using deep eutectic solvents and analytical methods for the isolation and characterization of natural bioactive compounds from plant material. Plants. 9(11). 1428. doi: https://doi.org/10.3390/plants9111428.
García-Roldán A., Piriou L., & Jauregi P. 2023. Natural deep eutectic solvents as a green extraction of polyphenols from spent coffee ground with enhanced bioactivities. Front. Plant Sci. 13. 1072592. doi: https://doi.org/10.3389/fpls.2022.1072592.
Maimulyanti A., Nurhidayati I., Mellisani B., Amelia R.P.F., Puspita F., & Restu P.A. 2023. Development of natural deep eutectic solvent (NADES) based on choline chloride as a green solvent to extract phenolic compound from coffee husk waste. Arab. J. Chem., 13. 1072592 . doi: https://doi.org/10.1016/j.arabjc.2023.104634.
Momotko M., Łuczak J., Przyjazny A., & Boczkaj G. 2022. A natural deep eutectic solvent-protonated L-proline-xylitol-based stationary phase for gas chromatography. J. Chromatogr A. 1676. 463238. doi: https://doi.org/10.1016/j.chroma.2022.463238.
Hikmawanti N.P.E., Ramadon D., Jantan I., & Mun’im A. 2021. Natural deep eutectic solvents (Nades): Phytochemical extraction performance enhancer for pharmaceutical and nutraceutical product development. Plants. 10(10). 2091. doi: https://doi.org/10.3390/plants10102091.
Zhao Y., Wan H., Yang J., Huang Y., He Y., Wan H., & Li C. 2022. Ultrasound-assisted preparation of ‘Ready-to-use’ extracts from Radix Paeoniae Rubra with natural deep eutectic solvents and neuroprotectivity evaluation of the extracts against cerebral ischemic/ reperfusion injury. Ultrason. Sonochem. 84. 105968. doi: https://doi.org/10.1016/j.ultsonch.2022.105968.
Sakti A.S., Saputri F.C., & Mun’im A. 2019. Optimization of choline chloride-glycerol based natural deep eutectic solvent for extraction bioactive substances from Cinnamomum burmannii barks and Caesalpinia sappan heartwoods. Heliyon. 5(12). e02915. doi: https://doi.org/10.1016/j.heliyon.2019.e02915.
Grudniewska A., Pastyrczyk N. 2022. New insight for spent hops utilization: simultaneous extraction of protein and xanthohumol using deep eutectic solvents. Biomass Convers. Biorefinery . 13(16). 14975-14986. doi: https://doi.org/10.1007/s13399-022-03462-5.
Sapone V., Cicci A., Franceschi D., Vincenzi S., & Bravi M. 2020. Antioxidant extraction and bioactivity preservation from winery by-products by natural deep eutectic solvents (NaDES). Chem. Eng. Trans. 79. 157–162. doi: https://doi.org/10.3303/CET2079027.
Aduloju E.I., Yahaya N., Zain N.M., Kamaruddin M. A., & Abd Hamid, M.A. 2023. An overview on the use of deep eutectic solvents for green extraction of some selected bioactive compounds from natural matrices. Adv. J. Chem. Sect. A. 6(3). 253–300. doi: https://doi.org/10.22034/AJCA.2023.389403.1356.
Bosiljkov T., Dujmić F., Bubalo M.C., Hribar J., Vidrih R., Brnčić M., Zlatic E., Redovniković, I.R. & Jokić S. 2017. Natural deep eutectic solvents and ultrasound-assisted extraction: Green approaches for extraction of wine lees anthocyanins. Food Bioprod. Process. 102. 195-203. doi: https://doi.org/10.1016/j.fbp.2016.12.005.
Teslić N., Santos F., Oliveira F., Stupar A., Pojić M., Mandić A., Pavlić B., Kljakić A.C., Duarte, A.R.C., Paiva A. & Mišan A. 2022. Simultaneous hydrolysis of ellagitannins and extraction of ellagic acid from defatted raspberry seeds using natural deep eutectic solvents (NADES). Antioxidants. 11(2). 254. doi: https://doi.org/10.3390/antiox11020254.
Sarma P.P., Gurumayum N., Verma A.K., & Devi R. 2021. A pharmacological perspective of banana: Implications relating to therapeutic benefits and molecular docking. Food. Funct. 12(11). 4749–4767. doi: https://doi.org/10.1039/D1FO00477H.
Hou S. 2022. Genistein: Therapeutic and preventive effects, mechanisms, and clinical application in digestive tract tumor. Evidence-based Complement Altern. Med. 5957378. doi: https://doi.org/10.1155/2022/5957378.
Liu D., Zhen W., Yang Z., Carter J.D., Si H., Reynolds K.A. 2006. Genistein acutely stimulates insulin secretion in pancreatic β-cells through a cAMP-dependent protein kinase pathway. Diabetes. 55(4). 1043–1050. doi: https://doi.org/10.2337/diabetes.55.04.06.db05-1089.
Rijai L., Tang S.T., Priastomo M., Siska S., Indriyanti N., Ambarwati N.S.S., & Ahmad I. 2023. Microwave-assisted extraction of polyphenols from Eleutherine bulbosa Mill. Urb. bulbs using choline chloride-sorbitol based natural deep eutectic solvent. J. Appl. Pharm. Sci. 13(6). 217–224. doi: https://doi.org/10.7324/JAPS.2023.130962.
Palos-Hernández A., Fernández M.Y.G., Burrieza J.E., Pérez-Iglesias J.L. & González-Paramás, A.M. 2022. Obtaining green extracts rich in phenolic compounds from underexploited food by-products using natural deep eutectic solvents: Opportunities and challenges. Sustain. Chem. Pharm. 29. 100773. doi: https://doi.org/10.1016/j.scp.2022.100773.
Ahmad A.R., Mun'im A. & Elya B. 2012. Study of antioxidant activity with reduction of DPPH radical and xanthine oxidase inhibitor of the extract of Ruellia tuberosa Linn. leaf. Int. Res. J. Pharm. 3(11). 66–70.
Munim A., Ramadhan M.G., & Soemiati A. 2013. Screening of endophytic fungi from Cassia siamea lamk leaves as α-glucosidase inhibitor. Int. Res. J. Pharm. 4(5). 128–131.
Ahmad I., Pertiwi A.S., Kembaren Y.H., Rahman A. & Muna A. 2018. Application of natural deep eutectic solvent-based ultrasonic assisted extraction of total polyphenolic and caffeine content from Coffe Beans (Coffea Beans L.) for instant food products. J. Appl. Pharm. Sci. 8(8). 138-143. doi: https://doi.org/10.7324/JAPS.2018.8819.
Vasileva B., Staneva D., Grozdanova T., Petkov H., Trusheva B., Alipieva K., Popova M., Miloshev, G., Bankova V. & Georgieva M. 2022. Natural deep eutectic extracts of propolis, Sideritis scardica, and Plantago major reveal potential antiageing activity during yeast chronological lifespan. Oxid. Med. Cell Longev. 2022(1). 8368717. doi: https://doi.org/10.1155/2022/8368717.
Hasanah S.U., Sukrasno S., & Hartati R. 2020. Perbandingan kandungan genistein pada berbagai varietas kedelai (Glycine max) di Indonesia. J. Penelit. Pertan. Tanam Pangan. 4(2). 113-118.
Sulistyowati E., Martono S., Riyanto S., & Lukitaningsing E. 2018. Analisis Daidzein dan Genistein pada Kedelai (Glycine max L. Merril) varietas anjasmoro, argomulyo dan dena 2 menggunakan metode KCKT. Media Farm. Indones. 13(1). 1299–1304.
Yunindarwati E., Ulfa E.U., Puspitasari E. & Hidayat M.A. 2017. Penentuan kadar genistein dan aktivitas hambatan tirosinase kedelai (Glycine max) terfermentasi Aspergillus oryzae. J. Ilmu Kefarmasian Indones. 14(1). 1-7.
Essa A.F., Teleb M., El-Kersh D.M., El Gendy A.E.N.G., Elshamy A.I., & Farag M.A.. 2023 Natural acylated flavonoids: their chemistry and biological merits in context to molecular docking studies. Phytochem. Rev. 22(6). 1469-1508. doi: https://doi.org/10.1007/s11101-022-09840-1.
Mahnashi M.H., Alyami B.A., Alqahtani Y.S., Jan M.S., Rashid U., Sadiq A., & Alqarni A.O. 2021. Phytochemical profiling of bioactive compounds, anti-inflammatory and analgesic potentials of Habenaria digitata Lindl.: Molecular docking based synergistic effect of the identified compounds. J. Ethnopharmacol. 273. 113976. doi: https://doi.org/10.1016/j.jep.2021.113976.
Mahnashi M.H., Alqahtani Y.S., Alyami B.A., Alqarni A.O., Alqahl SA, Ullah F., Sadiq A., Zeb A., Ghufran M., Kuraev A. & Nawaz A. 2022. HPLC-DAD phenolics analysis, α-glucosidase, α-amylase inhibitory, molecular docking and nutritional profiles of Persicaria hydropiper L. BMC Complement. Med. Ther. 22(1). 1–20. doi: https://doi.org/10.1186/s12906-022-03510-7.
Paşayeva L., Fatullayev H., Celik I., Unal G., Bozkurt N.M., Tugay O. & Abdellattif M.H. 2022. Evaluation of the chemical composition, antioxidant and antidiabetic activity of rhaponticoides iconiensis flowers: Effects on key enzymes linked to type 2 diabetes in vitro, in silico and on alloxan-induced diabetic rats in vivo. Antioxidants 11(11). 2284. doi: https://doi.org/10.3390/antiox11112284.
Niu Y., Cui W., Liu R., Wang S., Ke H., Lei X. & Chen L. 2022. Structural mechanism of SGLT1 inhibitors. Nat. Commun. 13(1). 6440. doi: https://doi.org/10.1038/s41467-022-33421-7.
Kumar M., Suhag R., Hasan M., Dhumal S., Radha, Pandiselvam R., Senapathy M., Sampathrajan V., Punia S., Sayed A.A. & Singh S. 2023. Black soybean (Glycine max (L.) Merr.): paving the way toward new nutraceutical. Crit. Rev. Food. Sci. Nutr. 63(23). 6208–6234. doi: https://doi.org/10.1080/10408398.2022.2029825.
Walker J.M. 2013. Computational Toxicology. New York, Heidelberg, Dordrecht, London: Springer. p. 642.
Katowah D.F., Hassaneen H.M. & Farghaly T.A., 2022. Novel Spiro-pyrrolizidine-oxindole and Spiropyrrolidine-Oxindoles: Green synthesis under classical, ultrasonic, and microwave conditions and molecular docking simulation for antitumor and type 2 diabetes. Arab. J. Chem. 15(7). 103930. doi: https://doi.org/10.1016/j.arabjc.2022.103930.
Fitri R.A., Lestari T.A., Sari Y., Sutriyo S. & Mun’Im A. 2020. Freeze drying of natural deep eutectic solvent (NADES) extract of green coffee bean (Coffea canephora Pierre ex A. Froehner). J. Res. Pharm. 24(2). 225–232. doi: https://doi.org/10.35333/jrp.2020.139.
Cannavacciuolo C., Pagliari S., Frigerio J., Giustra C.M., Labra M. and Campone L. 2022. Natural deep eutectic solvents (NADESs) combined with sustainable extraction techniques: a review of the green chemistry approach in food analysis. Foods. 12(1). 56. doi: https://doi.org/10.3390/foods12010056.
Popovic B.M., Micic N., Potkonjak A., Blagojevic B., Pavlovic K., Milanov D. & Juric T. 2022. Novel extraction of polyphenols from sour cherry pomace using natural deep eutectic solvents– Ultrafast microwave-assisted NADES preparation and extraction. Food Chem. 366. 130562. doi: https://doi.org/10.1016/j.foodchem.2021.130562.
Ozkan G. 2024. Valorization of artichoke outer petals by using ultrasound‐assisted extraction and natural deep eutectic solvents (NADES) for the recovery of phenolic compounds. J. Sci. Food Agric. 104(5). 2744-2749. doi: https://doi.org/10.1002/jsfa.13158.
Grønlien K.G., Pedersen M.E., & Tønnesen H.H. 2020. A natural deep eutectic solvent (NADES) as potential excipient in collagen-based products. Int. J. Biol. Macromol. 156. 394–402. doi: https://doi.org/10.1016/j.ijbiomac.2020.04.026.
Mehariya S., Fratini F., Lavecchia R., & Zuorro A. 2021. Green extraction of value-added compounds form microalgae: A short review on natural deep eutectic solvents (NaDES) and related pre-treatments. J. Environ. Chem. Eng. 9(5). 105989. doi: https://doi.org/10.1016/j.jece.2021.105989.
Musoev A., Numonov S., You Z. & Gao H. 2019. Discovery of novel DPP-IV inhibitors as potential candidates for the treatment of type 2 diabetes mellitus predicted by 3D QSAR pharmacophore models, molecular docking and de novo evolution. Molecules. 24(16). 2870. doi: https://doi.org/10.3390/molecules24162870.
Waskitha S.S.W., Istyastono E.P., & Riswanto F.D.O. 2023. Molecular Docking Study of Caffeic Acid as Acetylcholinesterase Inhibitor. J. Food Pharm. Sci. 11(3). 867-873. doi: https://doi.org/10.22146/jfps.7665.
Ajeet, Kumar A. & Mishra A.K. 2018. Design, molecular docking, synthesis, characterization, biological activity evaluation (against MES model), in-silico biological activity spectrum (PASS analysis), toxicological and predicted oral rat LD 50 studies of novel sulphonamide derivatives. Front. Biol. 13(6). 425-451. doi: https://doi.org/10.1007/s11515-018-1512-4.
Kaladhar D.S., Yarla N.S. & Anusha N. 2013. Functional analysis and molecular docking studies of medicinal compounds for AchE and BchE in Alzheimer’s disease and type 2 diabetes mellitus. Aging Dis. 4(4). 186-200.
Mao J., Wang G., Yang L., Tan L., Tian C., Tang L., Fang L., Mu Z., Zhu Z. & Li Y. 2023. Combined network pharmacology and molecular docking to verify the treatment of type 2 diabetes with Pueraria lobata radix and Salviae miltiorrhizae radix. Comput. Math. Methods Med. 2023(1). 9150324. doi: https://doi.org/10.1155/2023/9150324.
Stalin A., Kandhasamy S., Kannan B.S., Verma R.S., Ignacimuthu S., Kim Y., Shao Q., Chen Y. & Palani P. 2020. Synthesis of a 1, 2, 3-bistriazole derivative of embelin and evaluation of its effect on high-fat diet fed-streptozotocin-induced type 2 diabetes in rats and molecular docking studies. Bioorg. Chem. 96. 103579. doi: https://doi.org/10.1016/j.bioorg.2020.103579.
Behera R.K., Barik M.R., & Aneesha A. 2012. Molecular level targeting of hepatic glucokinase by computational docking approach in the treatment of type 2 diabetes. J. Chem. Pharm Res. 4(8). 3849-3854.
Gnaneswari D., Divya D., Verma A., Shraddha S., Sharma S. & Mayandi D.G. 2022. Molecular docking of potential Indian medicinal plant compounds against dengue viral proteins. Indian J. Tradit. Knowl. 21(3). 537-544. doi: https://doi.org/10.56042/ijtk.v21i3.32900.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Indonesian Journal of Chemical Studies

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.