Identification of Tropical Marine Diatom Chaetoceros dayaensis CBO from Bokor Island through Morphology and Genetic Marker Analysis
DOI:
https://doi.org/10.55749/ijcs.v1i1.2Keywords:
DNA barcode, Genetic marker, SEM imaging, Tropical marine diatomAbstract
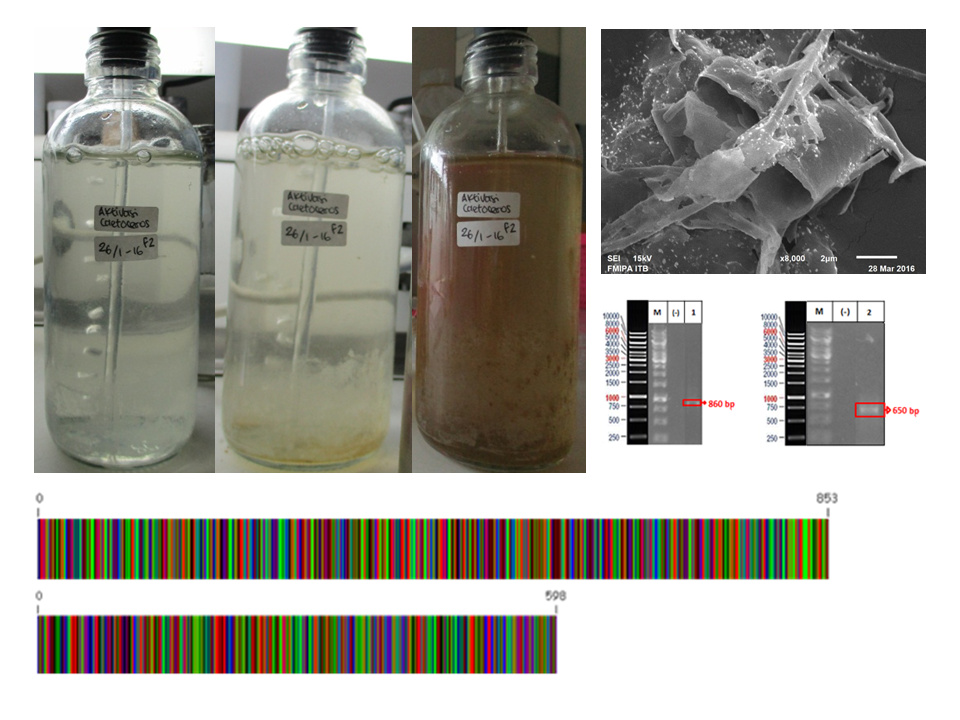
Microalgae are well-recognized by many researchers working on bioenergy resources due to its capability to produce high amounts of lipids as biodiesel precursors. Diatoms as a group of microalgae that specifically categorized by their silica valve could be more attractive to the researchers because in addition to high-lipid content, their silica features can be also utilized for wider applications, or used for diatoms bioproduct purification. Identification of some diatom species, such as the diatom genus Chaetoceros, as an initial step in diatom studies were difficult to accurately perform because of the unique morphology of silica frustules. Meanwhile, some candidate universal genetic markers of microalgae could also be used for the identification of diatoms with brittle frustules. In this research, we aimed to identify isolate CBO of tropical marine Chaetoceros CBO strain using morphological and genetic approaches. The isolate CBO of Chaetoceros sp. CBO was successfully obtained from sea region of Bokor Island, Kepulauan Seribu, Jakarta, and cultivated in our laboratory as culture stock for further use. Better resolution of the Chaetoceros CBO identification until species level was shown by genetic analysis toward two proposed gene markers for microalgae (rbcL-3P and V4 region of 18S rDNA) rather than by morphological. Specific DNA region for this specimen was found in V4 region of 18S rDNA genetic marker that could be recognized by MspA1I restriction enzyme. Hence, the RFLP (restriction fragment length polymorphism) method could be also used as an initial diagnostic tool for identification of this tropical marine Chaetoceros dayaensis CBO and for stock culture labelling purposes.
References
Barsanti, L., and Gualtieri, P. 2014. ALGAE: Anatomy, Biochemistry, and Biotechnology Second Edition. Boca Raton: CRC Press.
Street-Perrott, F.A., and Barker, P.A. 2008. Biogenic silica: a neglected component of the coupled global continental biogeochemical cycles of carbon and silicon. Earth Surf. Process. Landforms, 33, 1436-1457. doi: 10.1002/esp. https://doi.org/10.1002/esp.1712
Zeni, V., Baliota, G.V., Benelli, G., Canale, A., and Athanassiou, C.G. 2021. Diatomaceous earth for arthropod pest control: Back to the future. Molecules, 26(7487), 1-29 doi: 10.3390/molecules26247487. https://doi.org/10.3390/molecules26247487
Purnaningtyas, D. W., Aryantha, I. N. P., Nurachman, Z., & Rachmayanti, Y. (2020). Characterization of S-Acyltransferase Gene Fragment from an Isolate of Tropical Marine Microalgae Chlorella vulgaris CBI. IOP Conf. Ser. Earth Environ. Sci., 618(012036), 1-7. doi: 10.1088/1755-1315/618/1/012036. https://doi.org/10.1088/1755-1315/618/1/012036
Nurachman, Z., Brataningtyas, D.S., Hartati, and Panggabean, L. M. G. 2012. Oil from the tropical marine benthic-diatom navicula sp. Appl. Biochem. Biotechnol., 168(5), 1065-1075. doi: 10.1007/s12010-012-9841-2. https://doi.org/10.1007/s12010-012-9841-2
Nurachman, Z., Hartati, Anita, S., Anward, E.E., Novirani, G., Mangindaan, B., Gandasasmita, S., Syah, Y.M., Panggabean, L. M.G., and Suantika, G. 2012. Oil productivity of the tropical marine diatom Thalassiosira sp. Bioresour. Technol., 108, 240-244. doi: 10.1016/j.biortech.2011.12.082. https://doi.org/10.1016/j.biortech.2011.12.082
Damar, A., Colijn, F., Hesse, K.J., and Wardiatno, Y. 2012. The eutrophication states of Jakarta, Lampung and Semangka Bays: Nutrient and phytoplankton dynamics in Indonesian tropical waters. J. Trop. Biol. Conserv., 9(1), 61-81.
Rachmayanti, Y., Mwebaza, H., Radjawane, I. M., & Nurachman, Z. (2020). Tropical marine Navicula salinicola NBO: Morphology, genetic identification, and biochemical properties. IOP Conference Series: Earth and Environmental Science, 618(1), 1-11. doi: 10.1088/1755-1315/618/1/012033. https://doi.org/10.1088/1755-1315/618/1/012033
Hebert, P. D. N., Cywinska, A., Ball, S. L., and deWaard, J. R. 2003. Biological identifications through DNA barcodes. Proc. Biol. Sci. 270(1512), 313-321. doi: 10.1098/rspb.2002.2218. https://doi.org/10.1098/rspb.2002.2218
Hamsher, S. E., Evans, K. M., Mann, D. G., Poulíčková, A., and Saunders, G. W. 2011. Barcoding diatoms: exploring alternatives to COI-5P. Protist., 162(3), 405-422. doi: 10.1016/j.protis.2010.09.005. https://doi.org/10.1016/j.protis.2010.09.005
Zimmermann, J., Jahn, R., and Gemeinholzer, B. 2011. Barcoding diatoms: Evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Org. Divers. Evol., 11(3), 173-192. doi: 10.1007/s13127-011-0050-6. https://doi.org/10.1007/s13127-011-0050-6
Brodie, J., and Lewis, J. 2007. Unravelling the algae the past, present, and future of algal systematics. Boca Raton: CRC Press. https://doi.org/10.1201/9780849379901
Andersen, R.A. 2005. Algal Culturing Techniques. The Netherland: Elsevier Academic Press.
Romann, J., Valmalette, J.C., Chauton, M.S., Tranell, G., Einarsrud, M.A., and Vadstein, O. 2015. Wavelength and orientation dependent capture of light by diatom frustule nanostructures. Scientific Reports, 5(17403), 1-6. doi: 10.1038/srep17403. https://doi.org/10.1038/srep17403
Chen, X. G., Zhang, J., Huang, Y., and Hou, Y. P. 2013. Diatom taxa identification based on single-cell isolation and rDNA sequencing. Forensic Sci. Int. Genet. Suppl. Ser., 4(1), e308-e309. doi: 10.1016/j.fsigss.2013.10.157. https://doi.org/10.1016/j.fsigss.2013.10.157
Lang, I., and Kaczmarska, I. 2011. A protocol for a single-cell pcr of diatoms from fixed samples: Method validation using ditylum brightwellii (T. West) grunow. Diatom Research, 26(1), 43-49. doi: 10.1080/0269249X.2011.573703. https://doi.org/10.1080/0269249X.2011.573703
Levialdi Ghiron, J.H., Amato, A., Montresor, M., and Kooistra, W.H.C.F. 2008. Plastid Inheritance in the Planktonic Raphid Pennate Diatom Pseudo-nitzschia delicatissima (Bacillariophyceae). Protist., 159(1), 91-98. doi: 10.1016/j.protis.2007.06.002. https://doi.org/10.1016/j.protis.2007.06.002
Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis programfro Windows 95/98/NT. Nucleic Acids Symp. Ser., 41, 95-98.
Lee, S. D., Park, J. S., Yun, S. M., and Lee, J. H. (2014). Critical criteria for identification of the genus Chaetoceros (Bacillariophyta) based on setae ultrastructure. I. Subgenus Chaetoceros. Phycologia, 53(2), 174-187. doi: 10.2216/13-154.1. https://doi.org/10.2216/13-154.1
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2022 Indonesian Journal of Chemical Studies

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.